Neshyba & de Miguel, 2023¶
An Introduction to Statistical Mechanics¶
A brief introduction to StatMech¶
Statistical Mechanics is the name coined by Josiah Willard Gibbs back in 1884 to describe a set of methods people were inventing at the time, which were aimed at using microscopic properties of substances to explain their macroscopic properties (see https://en.wikipedia.org/wiki/Statistical_mechanics). Nowadays, those microscopic properties tend to be atomic-level properties that we can observe from spectroscopy experiments (like IR absorption experiments), or that we can predict from calculations (e.g., by solving Schrödinger's Equation). The macroscopic properties that StatMech tries to explain cover a huge range of possibilities, including heat capacities, thermoconductivity -- any bulk property you can measure. A great triumph of StatMech has been to beat a path from microscopic properties that one might get from Quantum Mechanics, to all the state functions of Thermodynamics -- including the internal energy, $U$, the Gibbs energy, the entropy, and the enthalpy.
But first we'll need to develop some foundational tools, starting most fundamentally with how molecules store energy.
How molecules store energy¶
You probably remember that molecules vibrate with certain frequencies that you can get from an IR absorption experiment. An example is shown in Fig. 1.
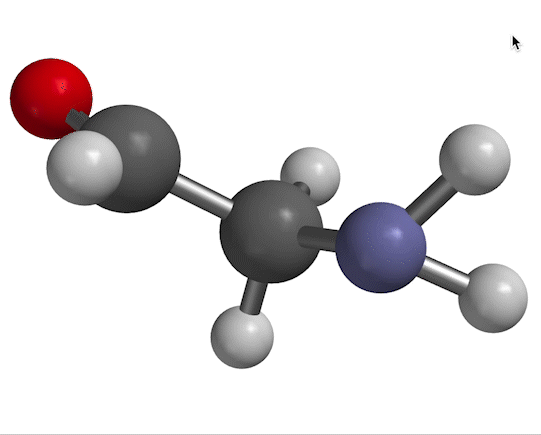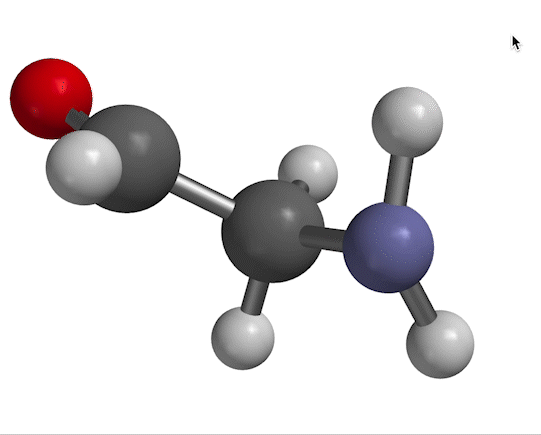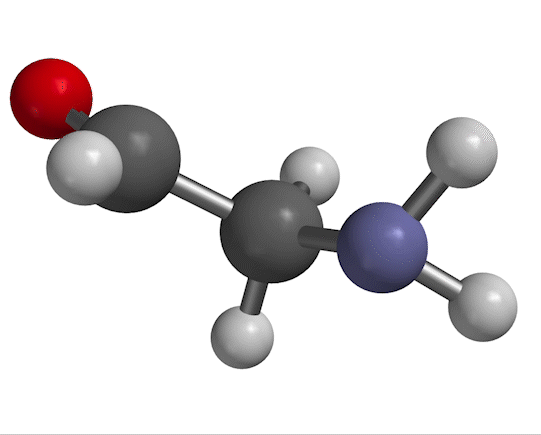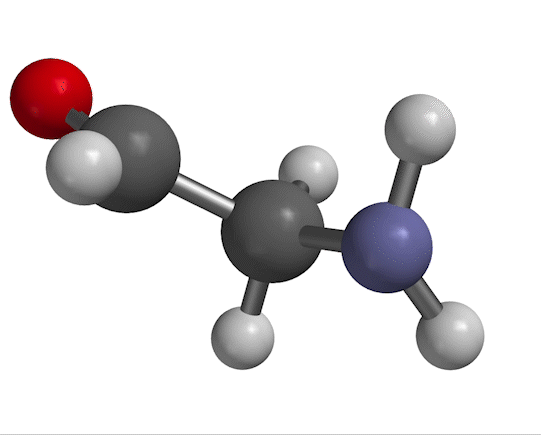 Figure 1. NH$_2$ scissors motion of 2-aminoacetaldehyde, with water solvent represented as a homogeneous dielectric medium.
Figure 1. NH$_2$ scissors motion of 2-aminoacetaldehyde, with water solvent represented as a homogeneous dielectric medium.
The motion we're looking at in Fig. 1 has a characteristic frequency of oscillation, which we typically designate with the symbol $\nu$. The same information appears in other forms, depending on the context. For example, in a typical IR spectrum, this motion would appear as a peak at wavenumber $\overline \nu$, given by
$$ \overline \nu = \nu/c \ \ \ \ (1) $$Some people prefer to think in terms of angular frequencies,
$$ \omega = 2 \pi \nu \ \ \ \ (2) $$These quantities tend to arise in quantum mechanics multipied by Planck's constant, where the math is just as simple one way or the other, since $h\nu=\hbar\omega$.
As for a quantum-mechanical analysis of this motion, a typical approximation (as you'll probably recall from previous work) is to describe the bending shown in Fig. 1 as a harmonic potential, ${1 \over 2}kx^2$ (where $x$ is the bending displacement you see in the figure). That information goes into Schrödinger's equation, which produces results like the ones shown in Fig. 2.
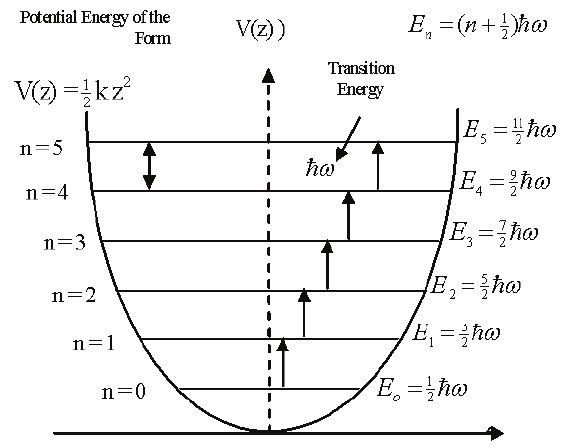 Figure 2. Vibrational energy levels.
Figure 2. Vibrational energy levels.
As Fig. 2 suggests, the eigenenergies associated with vibrational motion are given by
$$ E_n = (n + {1 \over 2}) h \nu = (n + {1 \over 2})\hbar \omega \ \ \ \ (3) $$where $n=0,1,...$ As you can see, the energy gaps between states are all the same,
$$ E_{gap} = \hbar \omega N_A \ \ \ \ (4) $$where we've tossed in Avogadro's number ($N_A$) to put these energies on a per-mole (rather than per-molecule) basis. So, if we wanted to focus on just the energy differences with respect to the ground state (which we do here), we could say
$$ \Delta E_n = n E_{gap} \ \ \ \ (5) $$The Partition Function and state functions derived from them¶
The foundational quantity of StatMech is the partition function, to which we'll give the symbol $Z$ (it also appears in the literature as $Q$). What exactly is $Z$? In physical terms, you can think of it as the number of quantum states available to a system at a given temperature.
An intuitive (but rough) estimate of $Z$ can be got from comparing the energy gap relative to the ground state of a system that we were just talking about, to the energy available in a typical intermolecular collision. That second quantity is given by ${3 \over 2}RT$, which (at room temperature) is about 4 kJ/mol (but of course gets bigger or smaller as the temperature goes up or down). Now, referring to Fig. 3, we can predict the following
- At sufficiently low temperatures, ${3 \over 2}RT << \Delta E_n$, so intermolecular collisions would hardly ever have enough energy to bump molecules out of the ground vibrational state. In that case, we'd expect $Z\approx 1$.
- As temperature goes up, at some point we'd find that the energy of intermolecular collisions (remember that's ${3 \over 2}RT$) would get close enough to $\Delta E_n$ that collisions could bump molecules in the ground vibrational state up one. We could expect $Z$ to be about equal to two in that case (and so on).
Figure 3 shows the results of a more quantitative treatment, assuming a vibrating molecule that has $E_{gap} \approx 1$ kJ/mol.
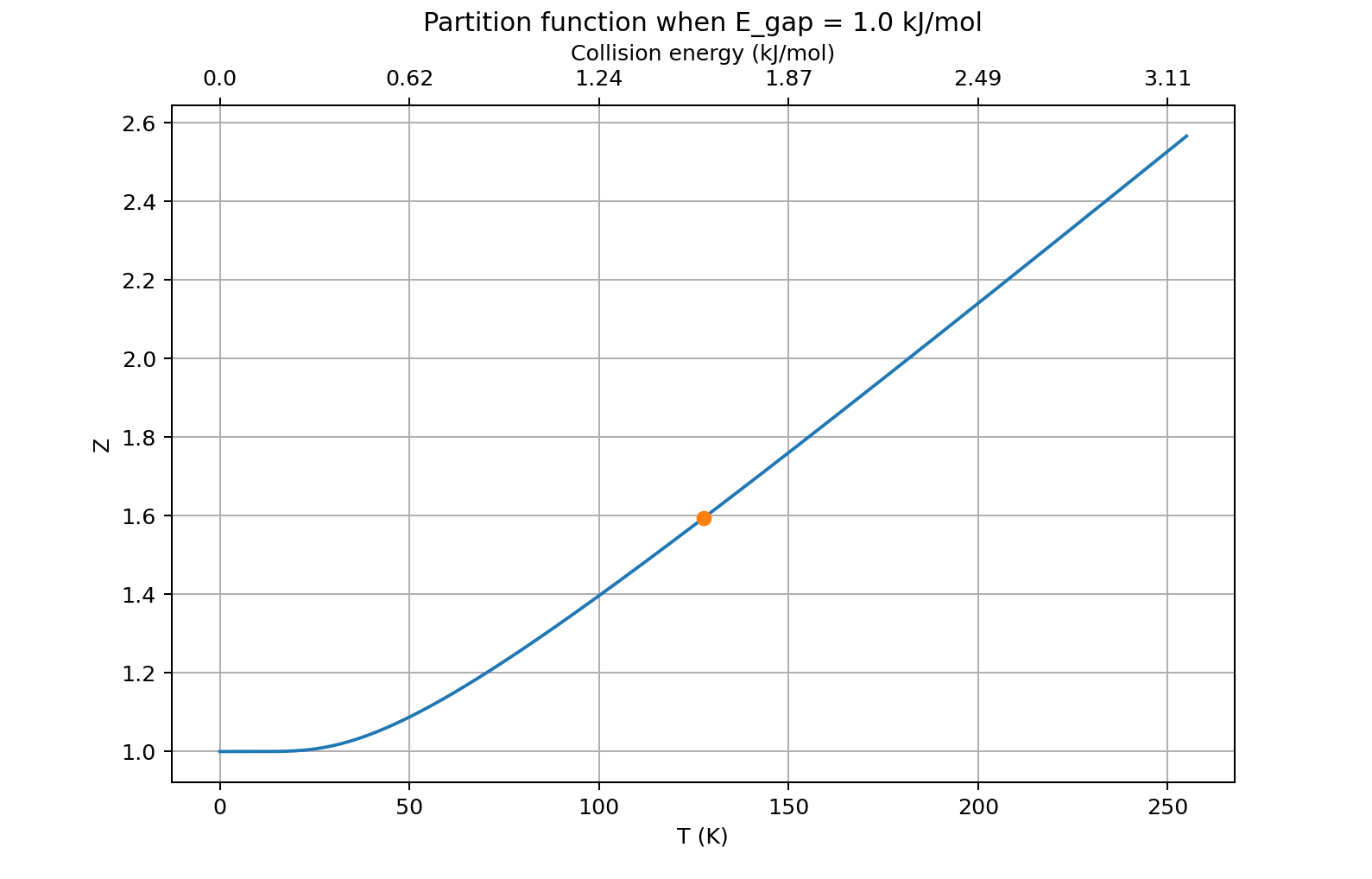 Figure 3. Partition function $Z(T)$ of a vibrating molecule having an energy spacing between vibrational levels of ~1 kJ/mol.
Figure 3. Partition function $Z(T)$ of a vibrating molecule having an energy spacing between vibrational levels of ~1 kJ/mol.
You can see that when the temperature is high enough that the energy of a typical collision is a little more than the energy spacing between vibrational levels (the marker), between one and two quantum states are available ($Z\approx 1.6$). And of course that number keeps going up as the temperature rises.
How do we compute partition functions such as the one in Fig. 3? It's surprisingly simple, actually:
$$ Z = \sum_{n} e^{-\Delta E_n / RT} \ \ \ \ (6) $$so as long as you know the energy levels $E_n$, you can find $Z$.
As advertised, the partition function can be used to derive expressions for all thermodynamic state functions. You can find a comprehensive list at https://en.wikipedia.org/wiki/Partition_function_(statistical_mechanics), but the one we'll need here is $U$,
$$ U = RT^2 {d \over dT} lnZ \ \ \ \ (7) $$Application to thermophoresis¶
The context we'll be considering is called thermophoresis, a phenomenon first documented in the mid-1800s, but of considerable contemporary interest too. But what is thermophoresis? As described in https://en.wikipedia.org/wiki/Thermophoresis,
"The phenomenon is observed at the scale of one millimeter or less. An example that may be observed by the naked eye with good lighting is when the hot rod of an electric heater is surrounded by tobacco smoke: the smoke goes away from the immediate vicinity of the hot rod. As the small particles of air nearest the hot rod are heated, they create a fast flow away from the rod, down the temperature gradient. They have acquired higher kinetic energy with their higher temperature. When they collide with the large, slower-moving particles of the tobacco smoke they push the latter away from the rod."
Here we'll consider the thermophoresis of a solute dissolved in a solvent, sandwiched between two thermal reservoirs, such as depicted in Fig. 4.
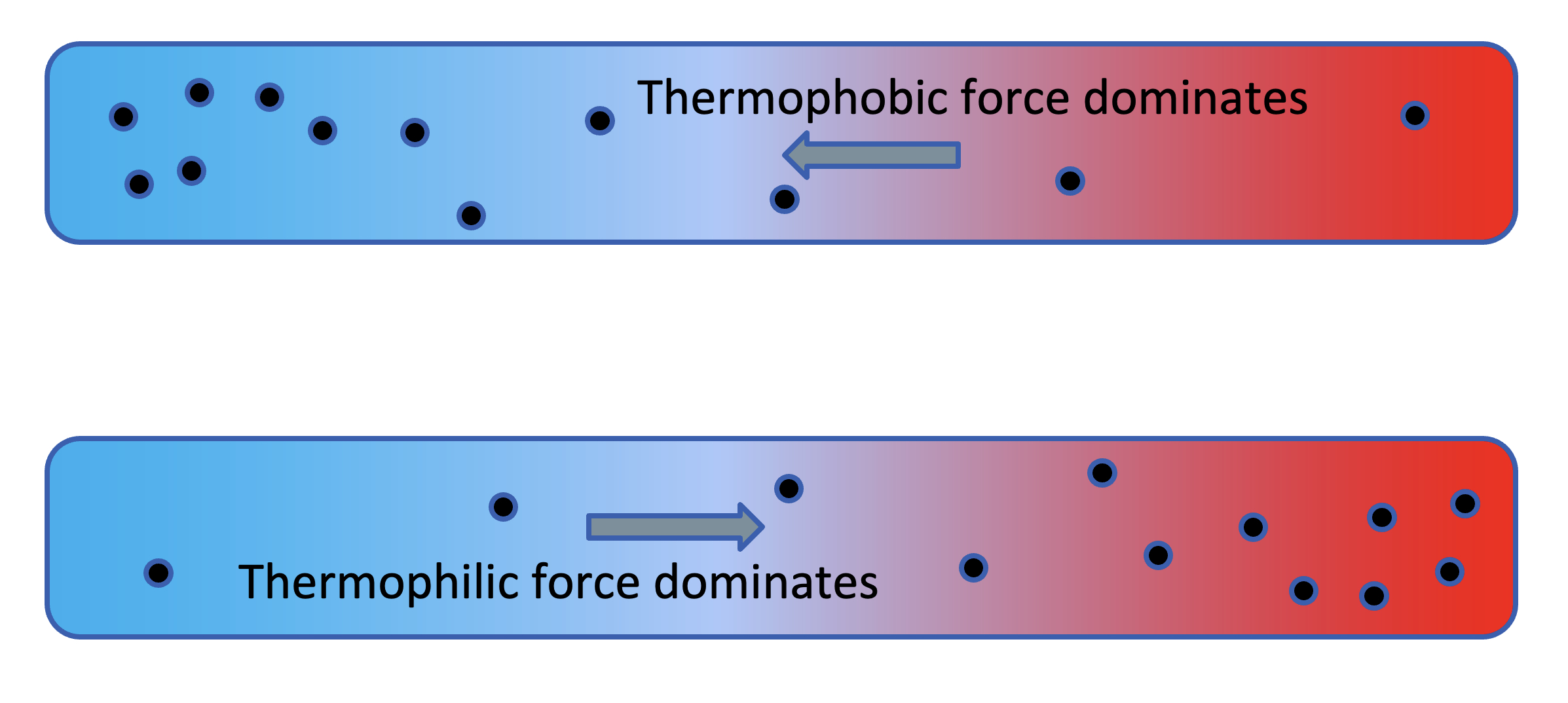 Figure 4. Evidence of thermophoretic forces.
Figure 4. Evidence of thermophoretic forces.
where the horizontal distance (call it $x$) spans a distance of (in this case) 0.1 cm. We're assuming an imposed, fixed temperature gradient as a function this distance,
$$ T(x) = T_{cold} + (T_{hot}-T_{cold}){x \over L} \ \ \ \ (8) $$Two possibilities are indicated in the figure. Thermophobic thermophoresis is where the solute moves away from the heat. Thermophilic thermophoresis is where the solute moves toward the heat.
Which is the correct picture? StatMech's answer is to construct the energy of the solute, $U(T)$, where the temperature runs from hot to cold across some distance $L$ (Eq. 8). Then, analogously to the way a ball moves downhill in a gravitational field (Fig. 5), to predict that solute molecules will also move "downhill," toward lower values of $U(T)$.
.a352190.gif) Figure 5. Ball rolling downhill.
Figure 5. Ball rolling downhill.
Temperature-independent vs temperature-dependent Hamiltonians¶
You can see from the foregoing that central to our analysis is the vibrational frequency (wavenumber) of our solute. How do we figure that out? Fortunately, Spartan can provide some answers, but there's a detail about this that we'll have to be cognizant of. It has to do with the temperature dependence of these vibrational motions.
$\overline \nu$ in the high-temperature limit. Because we think of high temperature as being a situation in which solvent water molecules form only short-lived hydrogen bonds to the solute, the solvent may as well be considered a homogeneous dielectric substance. Figure 1 was obtained in this way, in Spartan, by specifying "water" as a solvent. Therefore, we'll call the wavenumber we get for the scissor bend in this manner $\overline \nu_{HT}$.
If we are happy with this picture, the thermophoretic analysis is pretty straightforward. We form the partition function according to Eq. 6, using a constant $\overline \nu_{HT}$, and inspect the resulting $U(x)$ -- if we see that it slopes down to the left (the cold side), we can conclude that the solute is thermophobic. If we see that $U(x)$ slopes down to the right, it's thermophilic!
$\overline \nu$ in the low-temperature limit. On the other hand, if you inspect Fig. 6, you'll see that there's another way to find the frequency of vibration. When you set things up with solvent molecules explicitly this way, the result is considered a low-temperature limit. That's because now, the solvent water molecules can form long-lived hydrogen bonds to the solute. We'll call the wavenumber we get for the scissor bend in this case $\overline \nu_{LT}$.
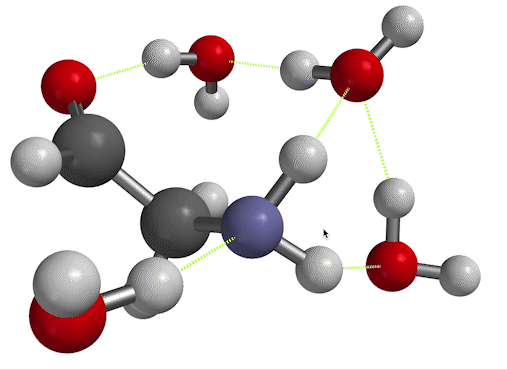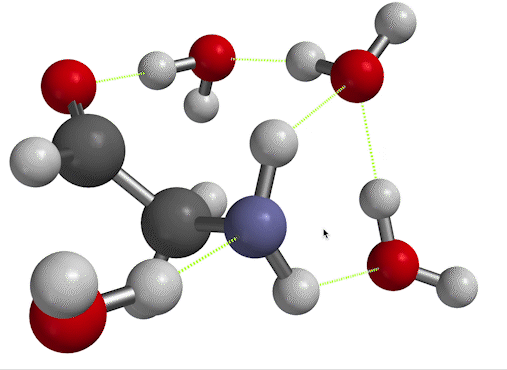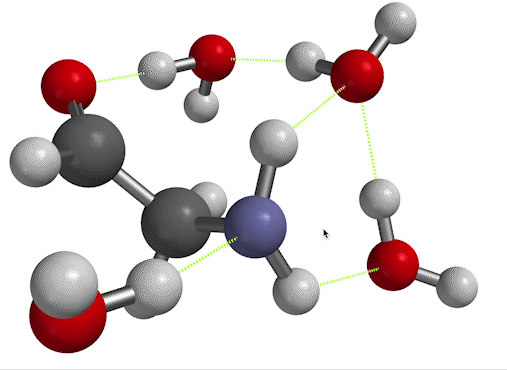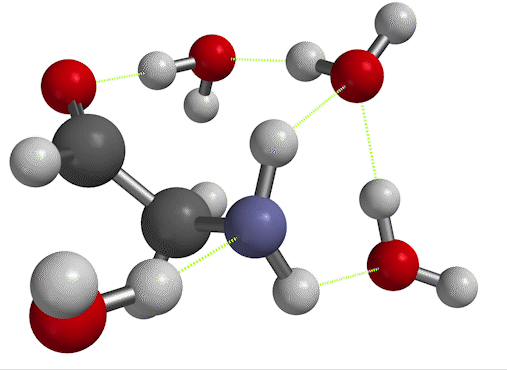 Figure 6. NH$_2$ scissors motion of 2-aminoacetaldehyde, in which the water solvent has been represented explicitly as well as implicitly.
Figure 6. NH$_2$ scissors motion of 2-aminoacetaldehyde, in which the water solvent has been represented explicitly as well as implicitly.
We'll also need a way to represent the transition from low temperature to high temperature. One possibility -- a sigmoidal transition -- is shown in Fig. 7.
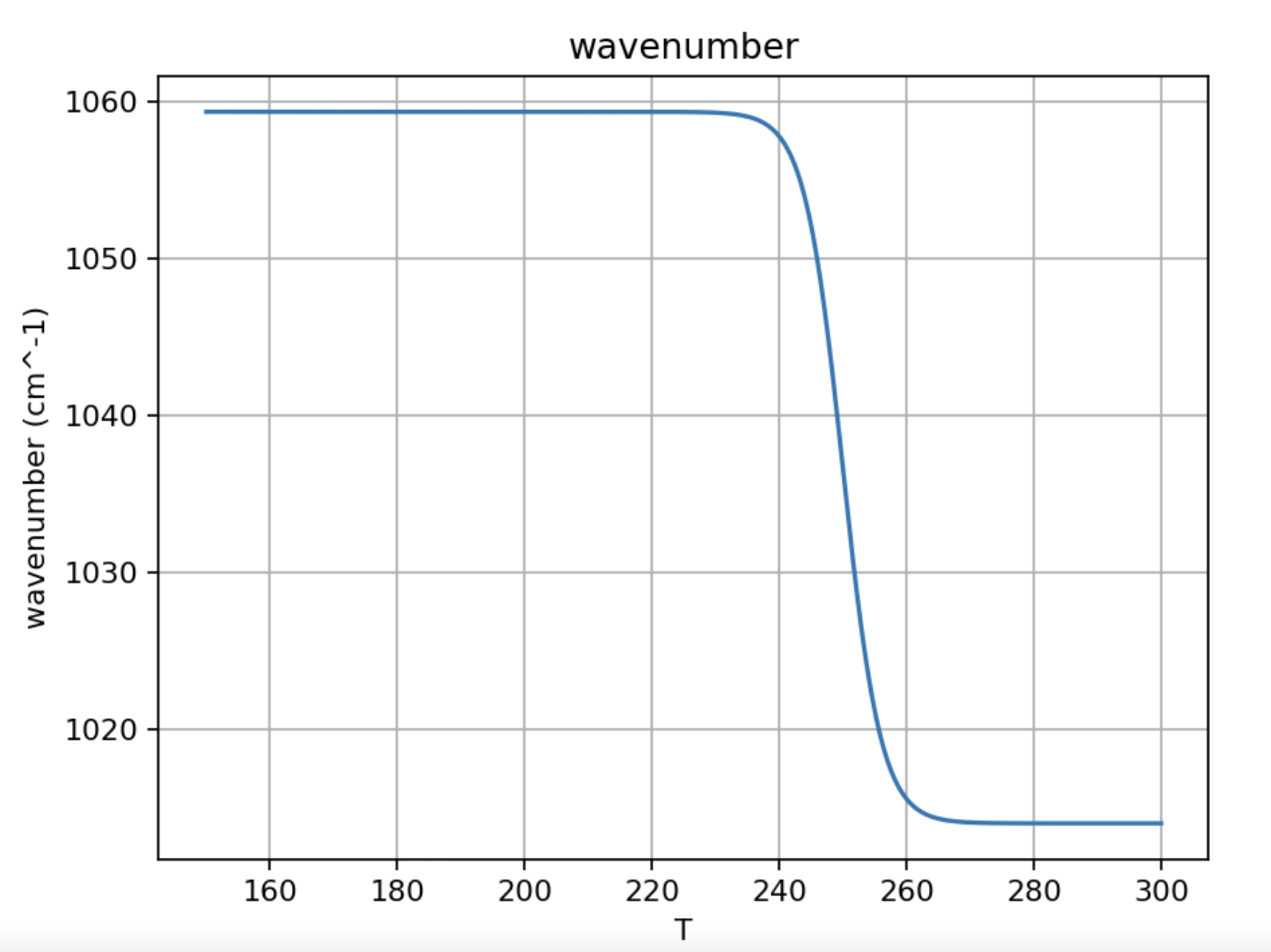 Figure 7. Sigmoidal temperature dependence of the vibrational frequency of an aqueous solute. The left-hand, low-temperature limit is designated $\overline \nu_{LT}$, the right-hand, high-temperature limit is designated $\overline \nu_{HT}$.
Figure 7. Sigmoidal temperature dependence of the vibrational frequency of an aqueous solute. The left-hand, low-temperature limit is designated $\overline \nu_{LT}$, the right-hand, high-temperature limit is designated $\overline \nu_{HT}$.
The thermophoretic analysis in this case here is almost the same as before, with the exception that because the frequencies of vibration are temperature-dependent, Eq. 6 becomes $Z = \sum_{n} e^{-\Delta E_n(T) / RT}$, where we've written $E_n=E_n(T)$ to emphasize this difference.
Idea of this CGI¶
The main idea of this CGI is to use Statistical Mechanics to predict the thermophoretic properties of a solute. There will be Spartan and Python components:
- In Spartan, you'll construct a solute molecule containing an amine group, and find the frequency of its scissors vibration assuming water as a solvent (first implicitly only, then surrounded by water molecules as an explicit solvent).
- In Python, you'll figure out whether the thermophoretic forces acting on a that solute are thermophyllic or thermophobic, first assuming temperature-independent vibrations (i.e., the high-temperature limit), and then assuming temperature-dependent vibrations (i.e., transitioning from the low-temperature limit to the high-temperature limit by means of a sigmoidal function).
Learning goals¶
The main learning goals of this exercise are
- I can construct state spaces needed for a StatMech problem.
- I can explain what the thermophoretic force is, including its thermophyilic an thermophobic versions.
- I can compute the energies, partition functions, and internal energy.
- I can interpret $U(T)$ in terms of thermophoretic forces.