# Reading in the emissions scenario file
filename = 'Scenario_2040_LTE4.hdf5'
time, eps, epsdictionary = CL.GetMyScenario(\
filename,reportflag=True,plotflag=True,epsdictionaryflag=True)
Here's the scenario summary:
{'LTE': 4,
'dataframe': time emissions
0 1750.000000 0.013363
1 1750.450450 0.013514
2 1750.900901 0.013667
3 1751.351351 0.013822
4 1751.801802 0.013978
.. ... ...
995 2198.198198 4.000000
996 2198.648649 4.000000
997 2199.099099 4.000000
998 2199.549550 4.000000
999 2200.000000 4.000000
[1000 rows x 2 columns],
'delta_t_trans': 20,
'emission units': 'GtC/year',
'eps_0': 11.3,
'k': 0.025,
'nsteps': 1000,
't_0': 2020,
't_peak': 2040,
't_start': 1750,
't_stop': 2200}
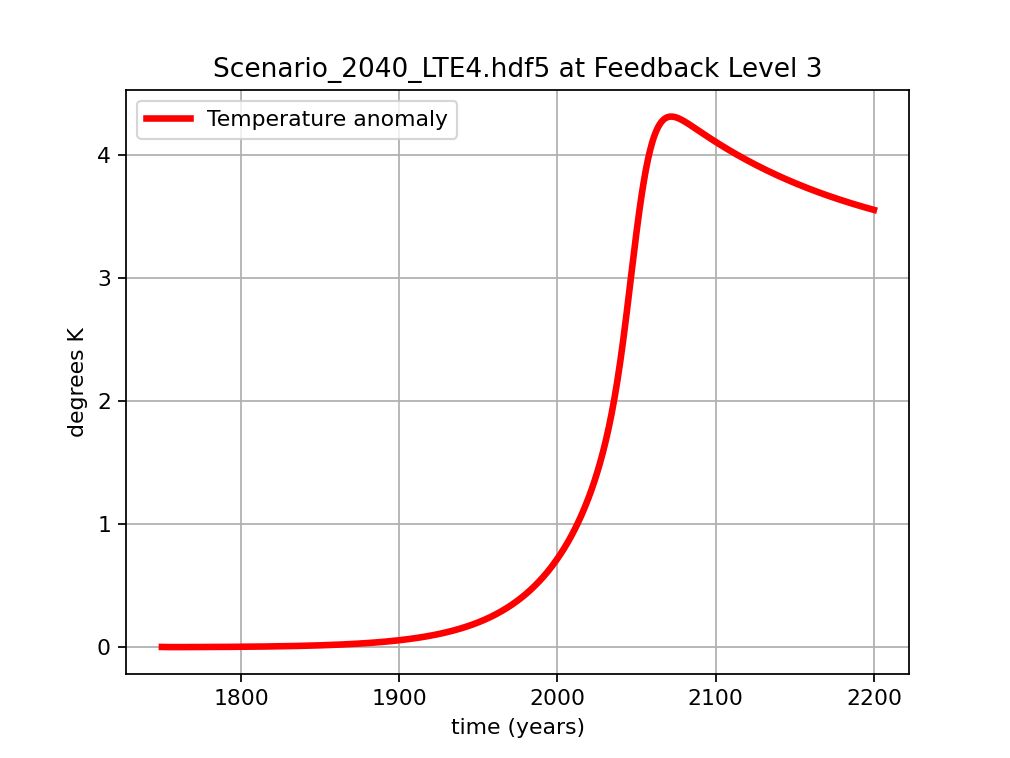 Sample temperature for Level 3 feedbacks.
Sample temperature for Level 3 feedbacks.