The Grinstead Lab Page
Puget Sound
My research has focused over the last several years on application of structural (NMR) and biophysical (UV/Vis and fluorescence spectroscopy, circular dichroism, and molecular modeling) techniques to biological systems. Specifically, I am interested in characterizing the structure, dynamics, thermodynamics and kinetics of protein-protein and protein-ligand interactions. As illustrated by the projects below, these studies can give data helpful in understanding basic biology or the biochemical basis for a disease.
Students interested in doing research with me may have the opportunity to use some of the above-mentioned techniques to characterize the structure and dynamics of ligands within the active site of protein molecules, and use the docking program HADDOCK (http://www.nmr.chem.uu.nl/haddock/) to use different types of spectroscopic and biochemical data to computationally dock protein-protein or protein-ligand complexes. To perform these experiments, it may be necessary to produce and purify recombinant proteins, chemically derivatize free sulfhydryl groups on the protein with paramagnetic labels, identify site(s) of derivatization using LC-MS, or assign complex NMR spectra. Of course, astute use of the relevant literature will focus the research direction, and will provide data for inclusion in the molecular modeling of complexes.
Cytochrome P450

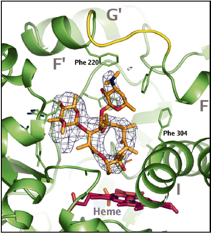
Structure of human P450 3A4 with erythromycin bound in the active site.